Following is a quick tour of the many displays and queries available through SyMAP.
Whole genome views
SyMAP can be used to query and view multiple synteny genome pairs at once (2-3 is best).
The synteny can be viewed from the
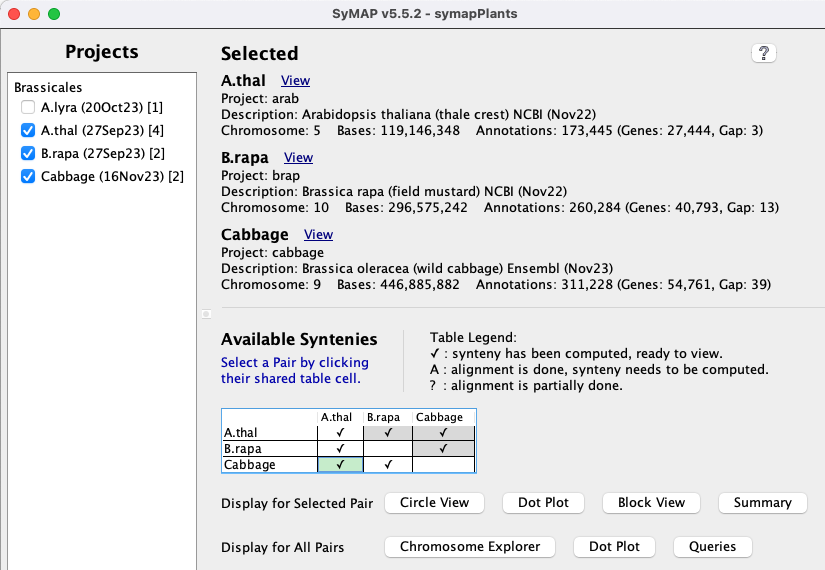
Project Manager.
All projects in the database will be listed on the left panel.
Selecting projects on the left panel shows them on the right panel.
A check mark in the Available Syntenies matrix indicates pairs that
have computed synteny that can be viewed.
These can be viewed from the lower multi-genome Display for All Pairs options.
By selecting a cell with synteny, the Display for Selected Pair buttons are activated, which
display synteny views for the two-genome.
The Project Manager
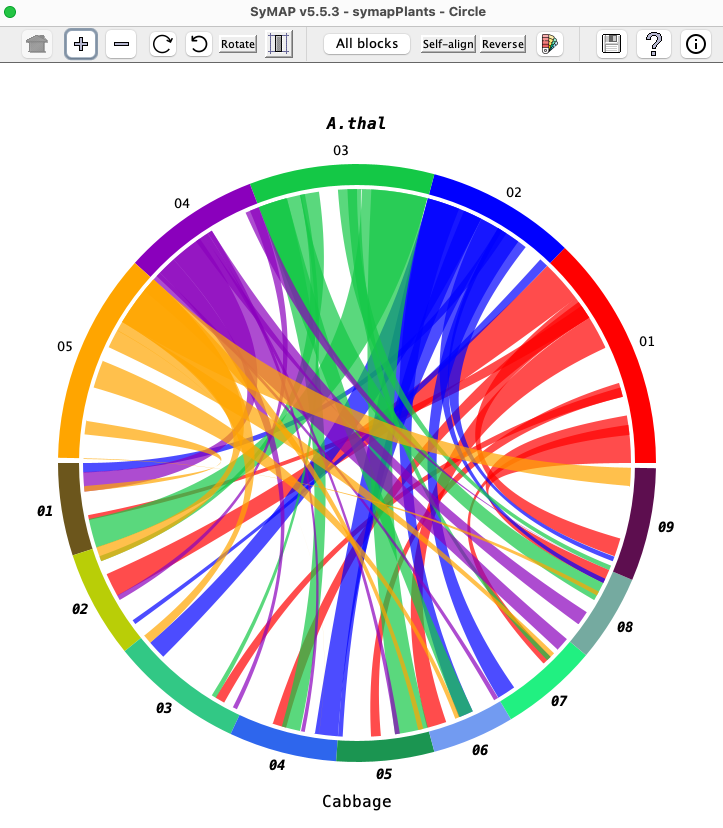
| The Circle view shows two genomes.
|
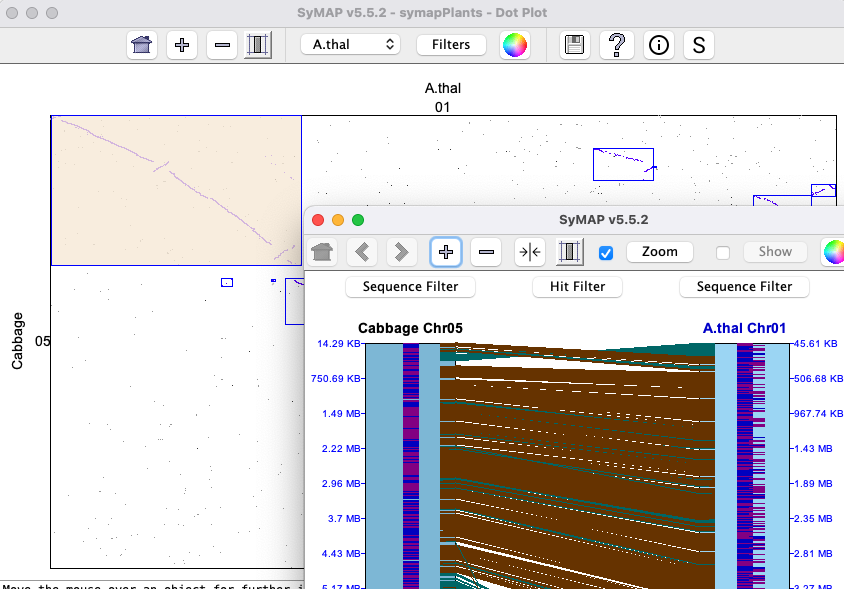
The Dot plot can show two or more genomes where the image on the lower left shows three.
A cell can be clicked to view a chromosome x chromosome Dot plot, from which a 2D
view can be displayed.
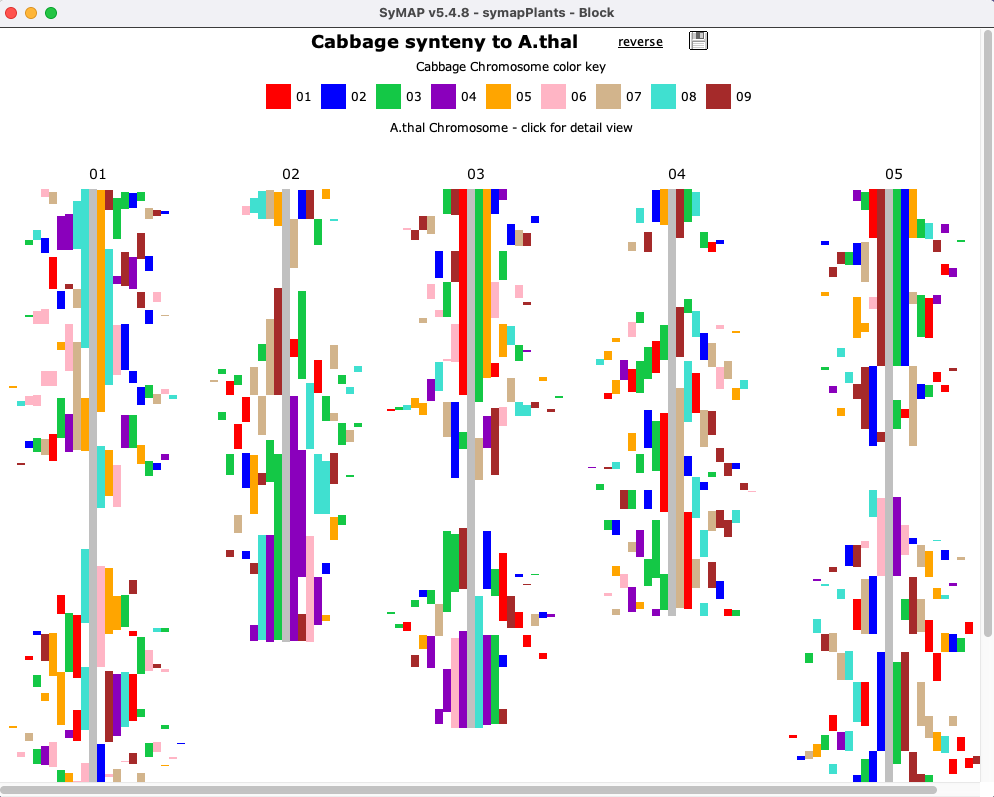
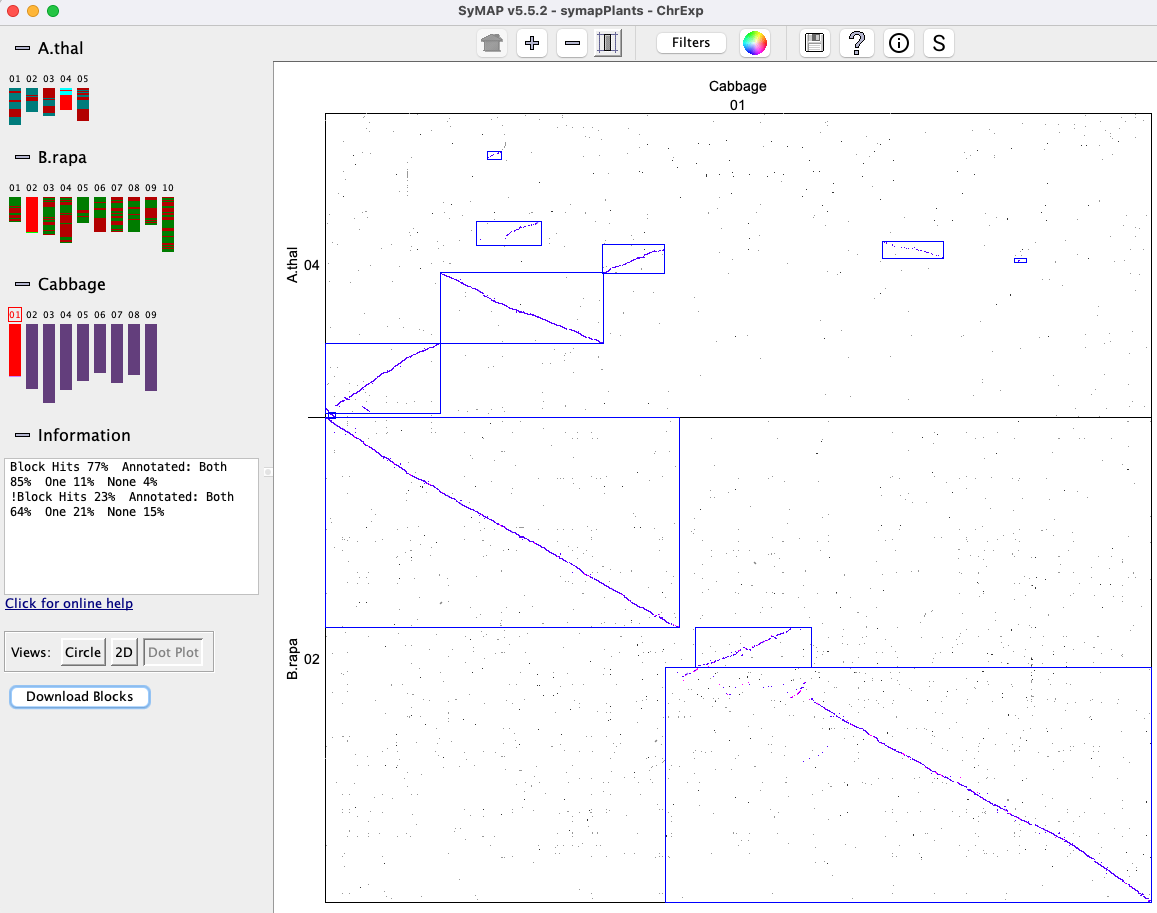
The Block view shows two genomes.
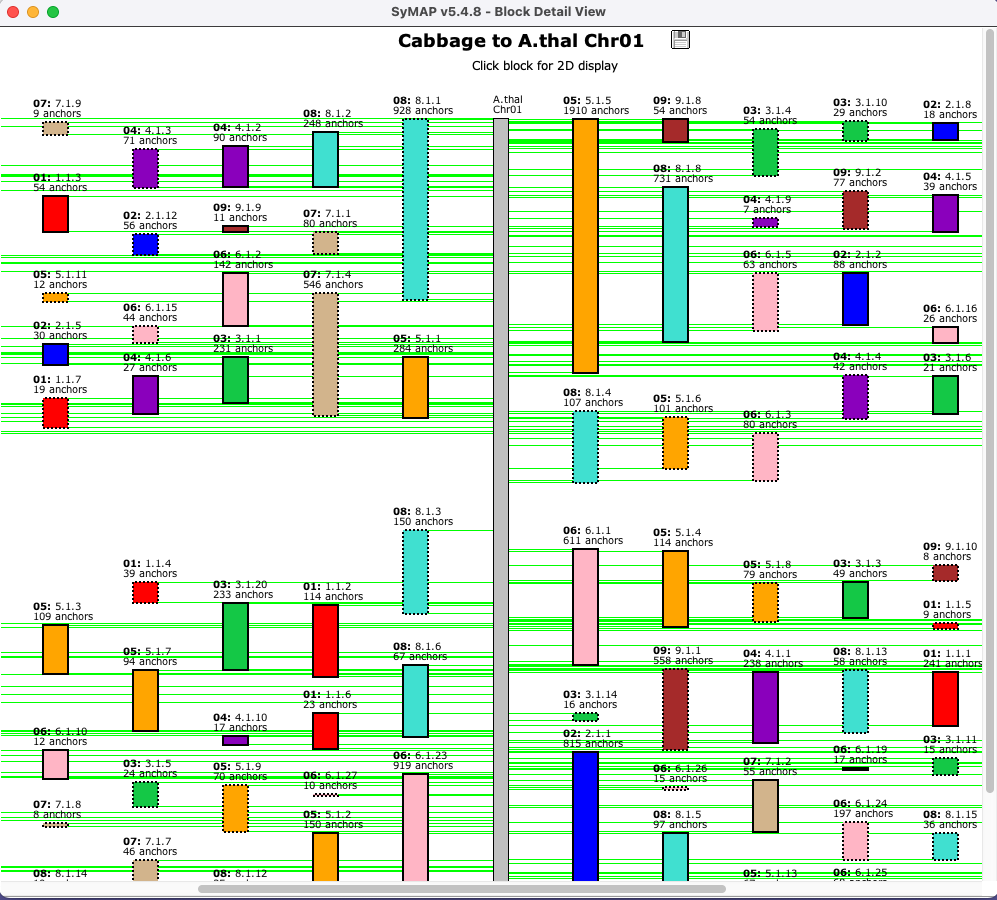
Clicking a chromosome shows a close-up of it, where a block can
be clicked to view the 2D display of it.
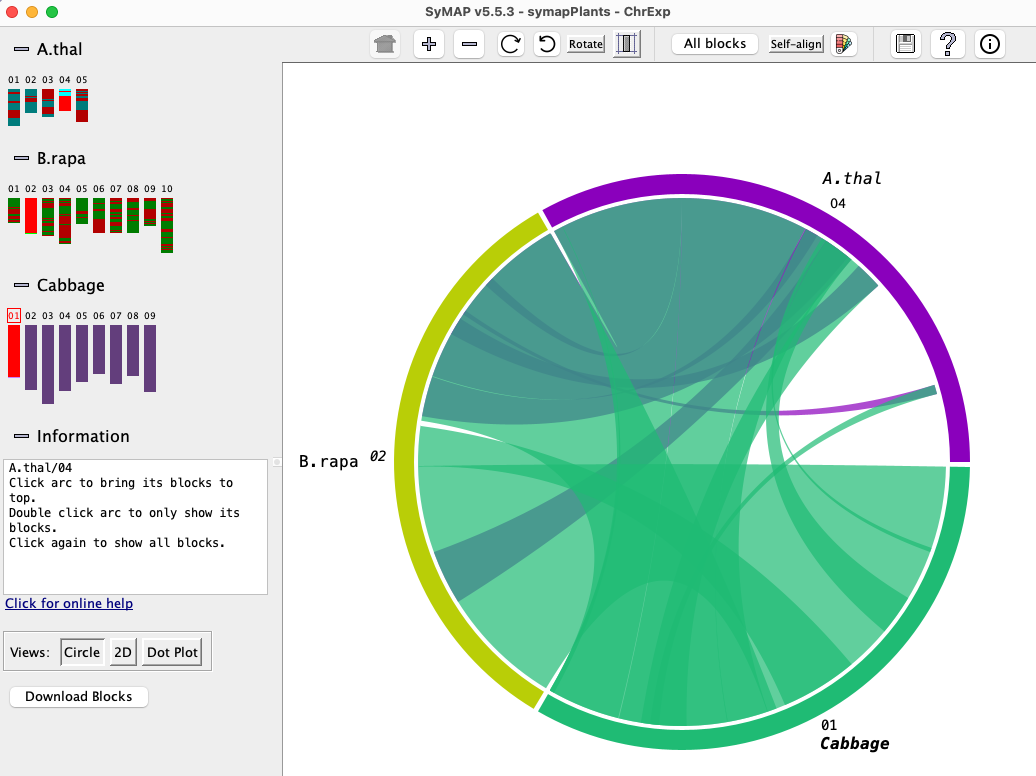
Chromosome Explorer
The
Chromosome Explorer provides
multi-chromosome views for
multi-genomes.
Multiple chromosomes can be selected from different species on the left-hand panel.
The default display is the Circle view shown. From here, one can select Dot Plot or 2D.
2D 3-Chr
|
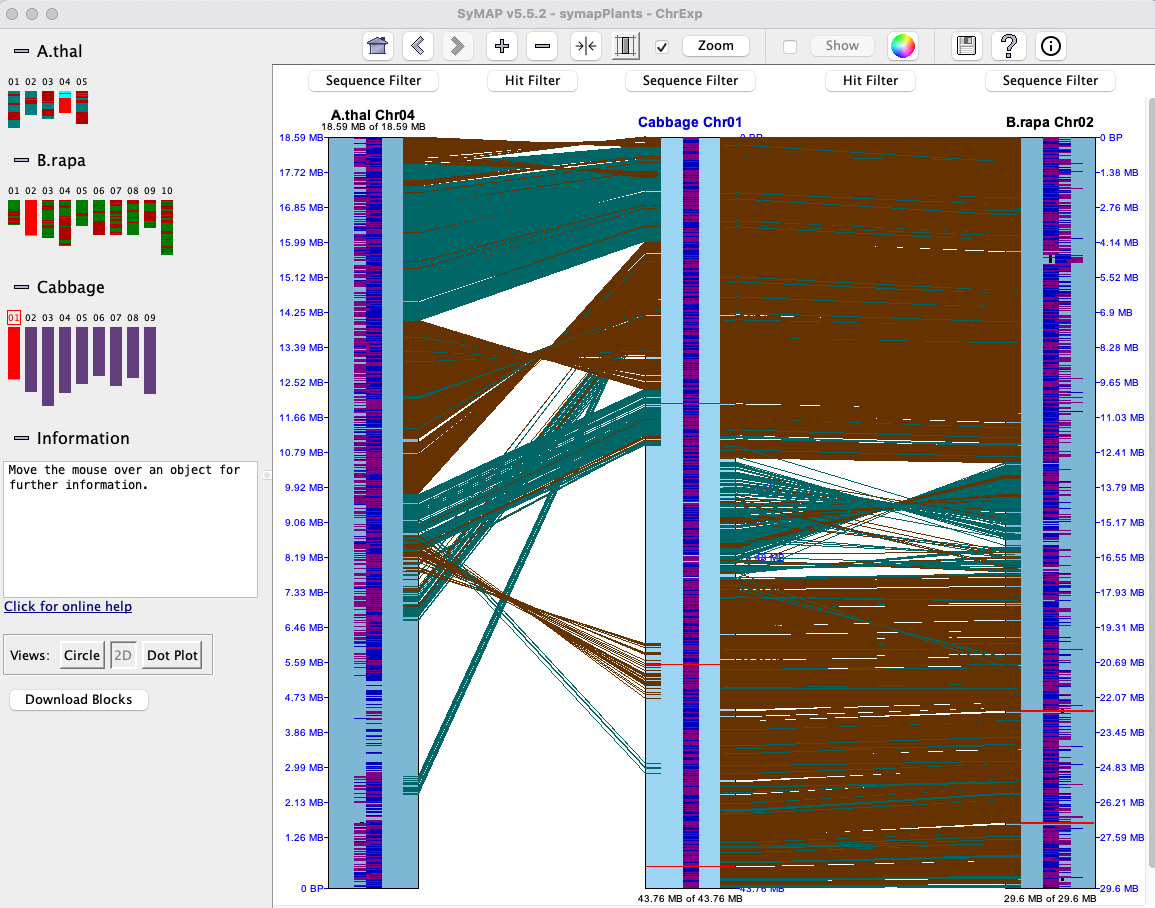
This is a three-chromosome view.
The dark blue and purple down the center of the chromosomes are annotated exons,
where blue is positive strand and purple is negative strand.
The brown and green lines between chromosomes are hit-wires connecting similar
regions on the two chromosomes. The brown are hit-wires with the same orientation on both
connecting chromosomes and the green are different orientation.
Only hits that are part of a synteny block are shown here.
| 
|
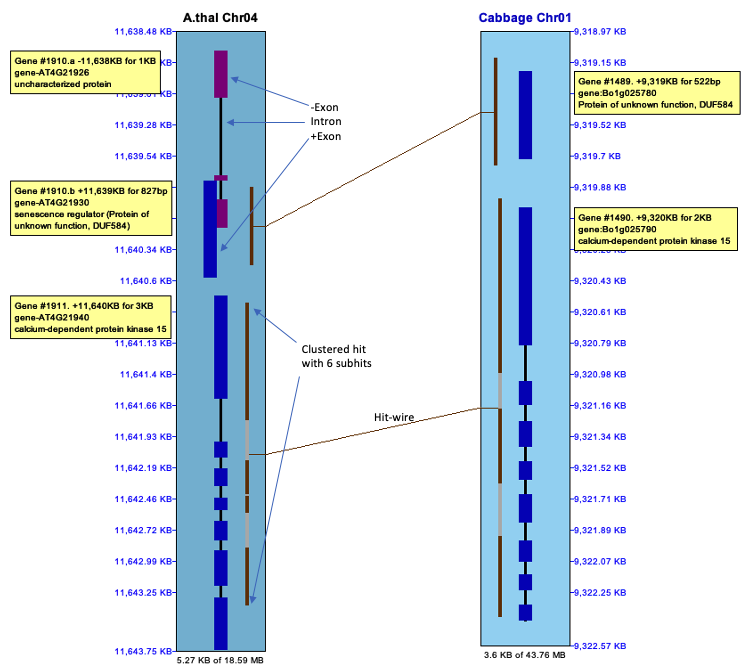
2D Closeup
|
The image on the right shows a smaller region with the Annotation option
is turned on.
A gene can be right clicked to view a popup of extended information,
including the exons and hits.
The hit-wire can be right clicked to view of popup of details of its information.
| 
|
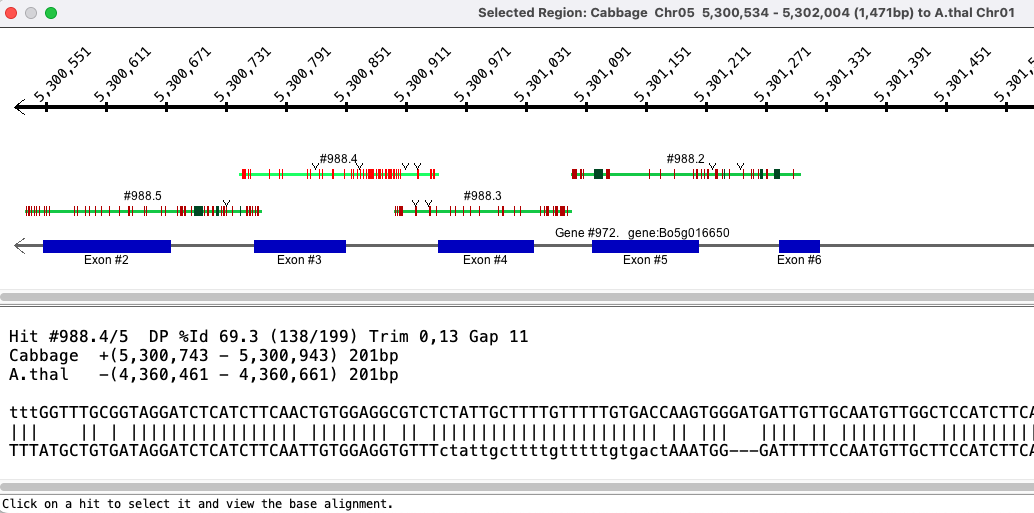
Base View
| It is possible to zoom in even further and see a basepair-alignment
of the hits in the region.
| 
|
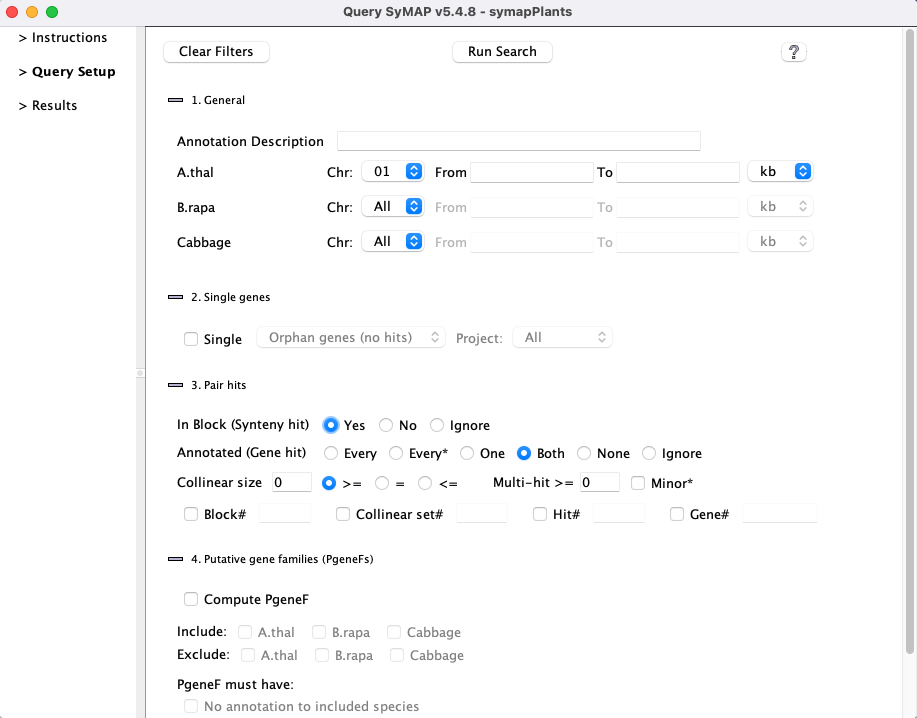
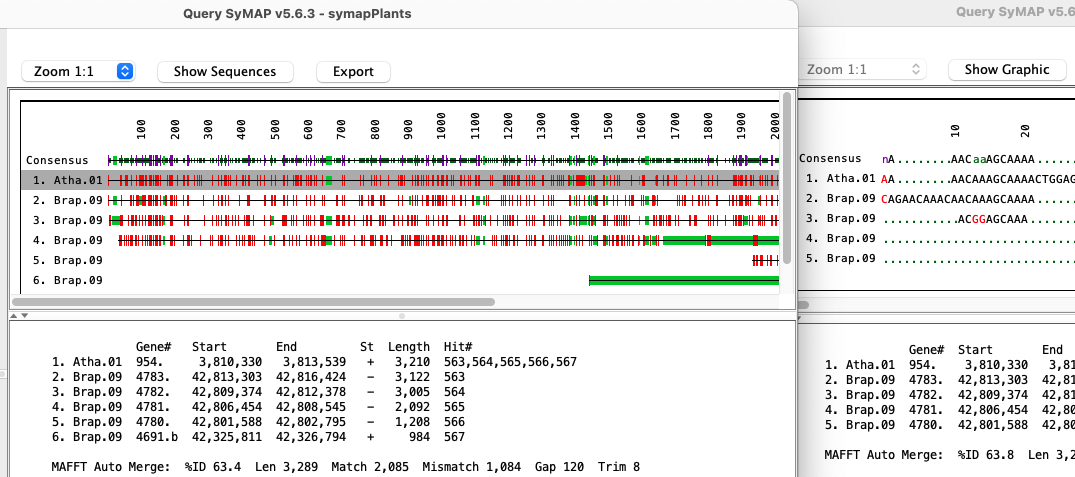
Query Interface
SyMAP includes powerful search features to help make the most of synteny information.
The query results table has full flexibility in column selection and ordering.
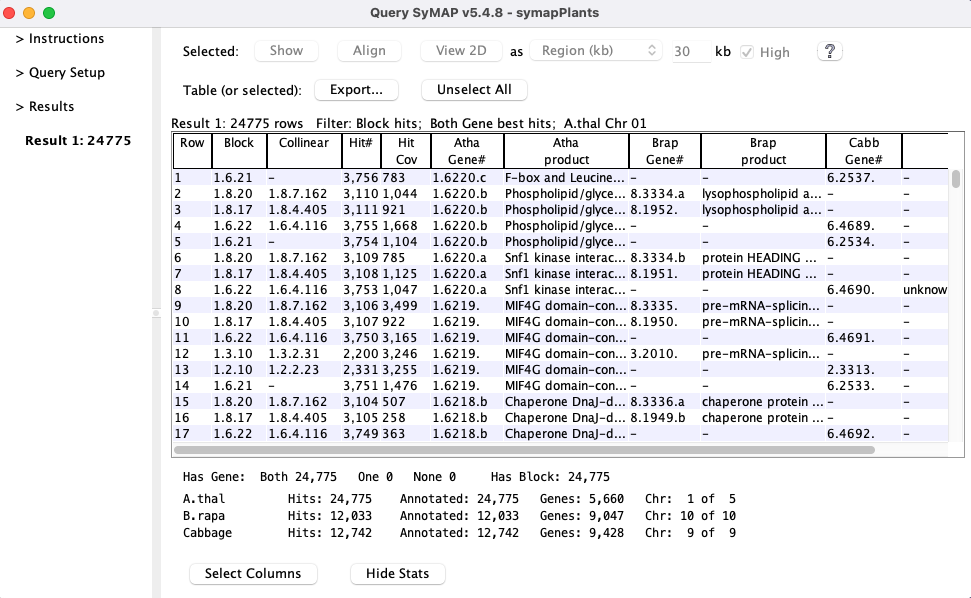
By selecting a one or more rows, the genes sequences can be aligned using
MUSCLE or MAFFT (
Refs6,7).
Email Comments To: cas1@arizona.edu