SyMAP ( Synteny Mapping and Analysis Program) is a software package for
detecting, displaying, and querying syntenic relationships between sequenced genomes.
- It is designed for divergent eukaryotic genomes (not bacteria).
- It can mask the non-genic sequence for gene synteny, or leave it un-masked for new gene discovery and whole genome synteny.
- It can align a draft genome to a fully sequenced genome (see Peach) and order the draft.
- The query and display is designed for the comparison of a few genomes at a time (i.e. 2-4).
Latest SyMAP release: v5.7.9c (4-Jan-2026), see Release Notes.
|
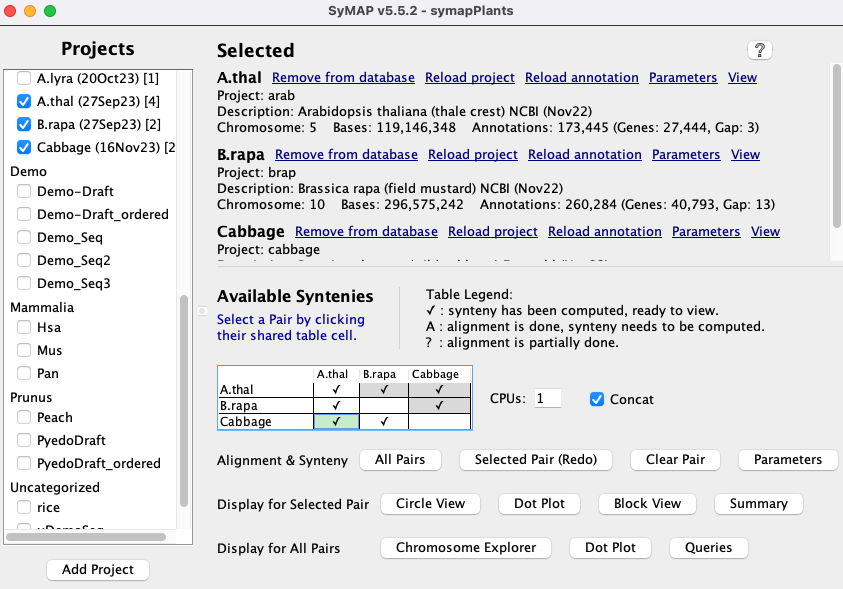
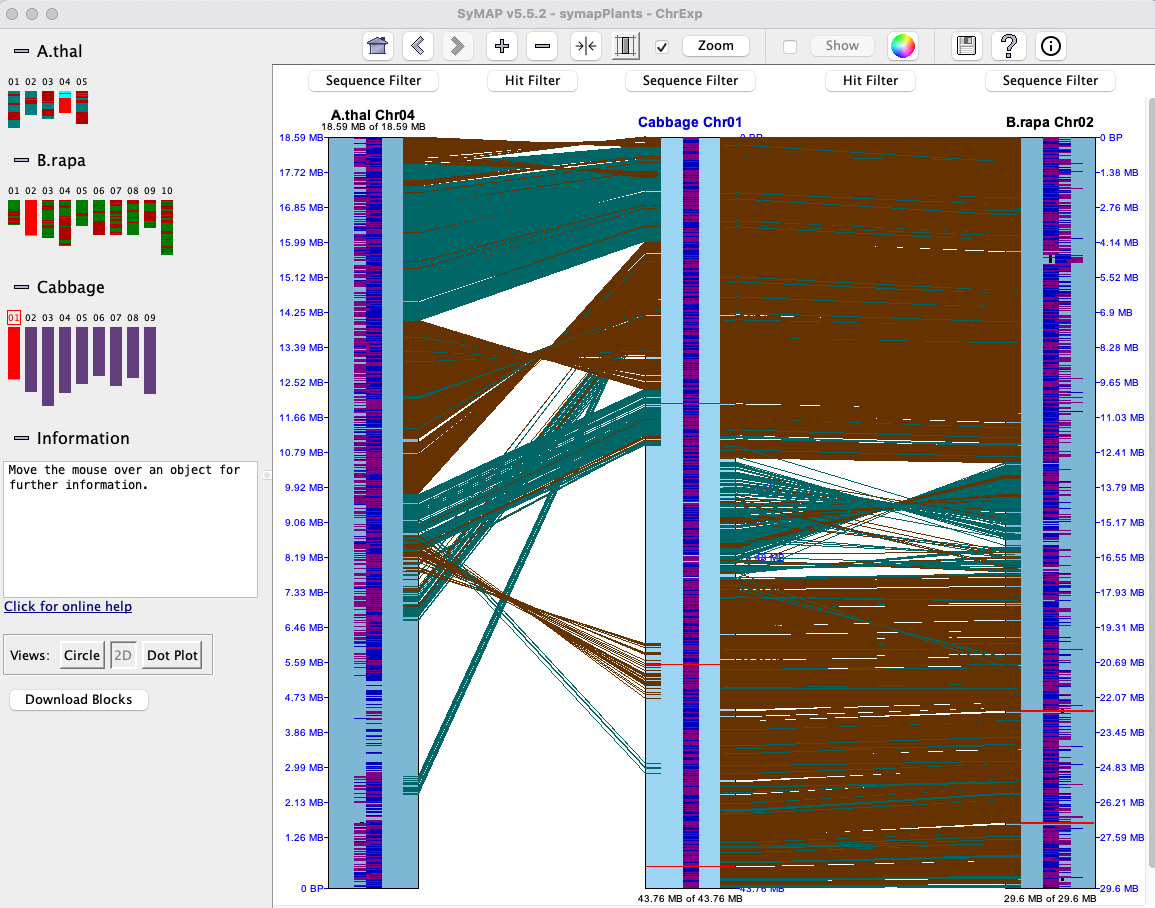
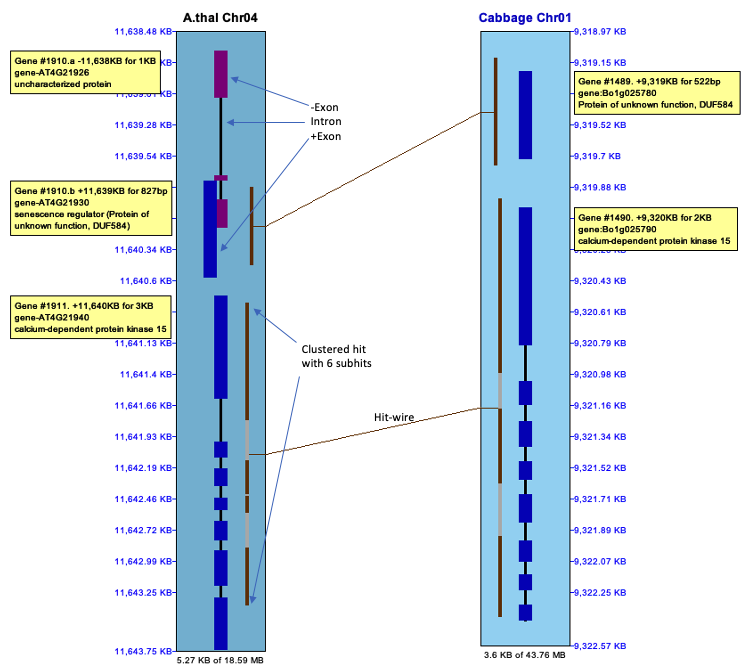
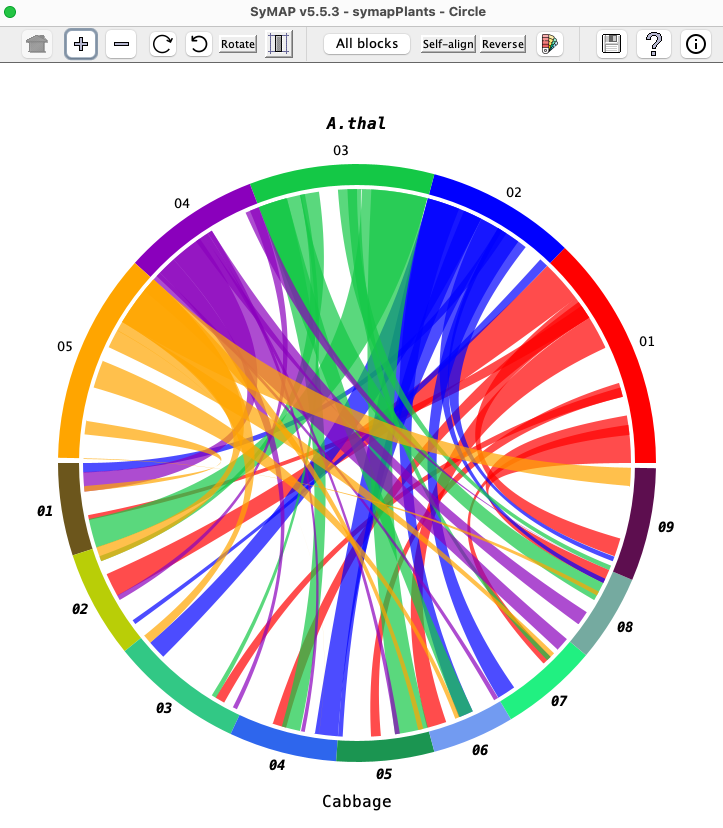
| Click an image to see the closeup.
|

| This project was partially supported by the National Research Initiative of USDA's National
Institute of Food and Agriculture, grant #0214643.
|  All upgrades since release 7-Nov-2016 have been developed by CAS without funding.
All upgrades since release 7-Nov-2016 have been developed by CAS without funding.
SyMAP was originally written for diverse plant genomes with short introns, but has been modified to work
for the long introns of mammalian genomes, and less diverse genomes.
|
Publications
C. Soderlund, M. Bomhoff, and W. Nelson (2011) SyMAP v3.4: a turnkey synteny system with
application to plant genomes Nucleic Acids Research 39(10):e68
Link
C. Soderlund, W. Nelson, A. Shoemaker and A. Paterson (2006) SyMAP: A system for
discovering and viewing syntenic regions of FPC maps. Genome Research 16:1159-1168.
Link
|